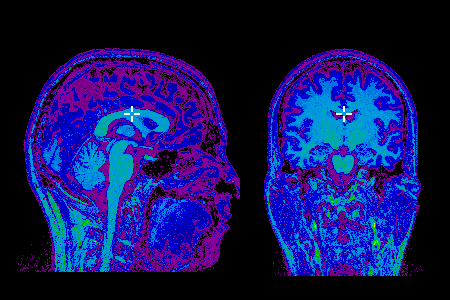
BrainBrowser is...

BrainBrowser is an open source JavaScript library exposing a set of web-based 3D visualization tools primarily targetting neuroimaging [Sherif et al. 2015]. Using open web-standard technologies, such as WebGL and HTML5, it allows for real-time manipulation and analysis of 3D imaging data through any modern web browser. The BrainBrowser Surface Viewer is a WebGL-based 3D viewer capable of displaying 3D surfaces in real-time and mapping various sorts of data to them. The BrainBrowser Volume Viewer is an HTML5 canvas-based viewer allowing slice-by-slice traversal of MINC, NIfTI and MGH/freesurfer volumetric data.
Try them out in the following sample applicatons:
BrainBrowser is developed and maintained as part of the CBRAIN project at the McGill Centre for Integrative Neuroscience. BrainBrowser was originally created by Nicolas Kassis and Tarek Sherif, then developed and maintained by Robert D. Vincent and Natacha Beck.
Downloads
You can prepare your MINC data for use in the Volume Viewer using either minc2volume-viewer.js for node.js or minc2volume-viewer.py for python (requires installation of the MINC tools).
Documentation
BrainBrowser API documentation
Examples of BrainBrowser usage can be found in the Surface Viewer and Volume Viewer demonstration code used for this site.
Other information:
Publication:Web Service
v0.3.4
Load a BrainBrowser Surface Viewer widget into your web page to view your own data! Simply make an HTTP request to the following URL:
https://brainbrowser.cbrain.mcgill.ca/surface-viewer-widget
and insert the returned HTML into your page. The Web Service will automatically load all required library files and launch the viewer. Optionally, you can provide a callback function to programatically control the viewer.
Try out some of the basic web service functionality in the BrainBrowser Web Service Demo.
Please note the Surface Viewer Web Service is still in its early stages of development. Performance is not guaranteed and the API may change rapidly. Please send any comments or feedback at cbrain-support.mni@mcgill.ca.